Caret 26
Done. 55,644 rows loaded




Modified: 13.04.15 09:34 Size: 2.09 MB


Pharma_discovery
Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label








Label














File Explorer












Sequence Miner - [Pharma_discovery]





Report Builder
Report Builder
Report Builder
Report Builder
Report Builder



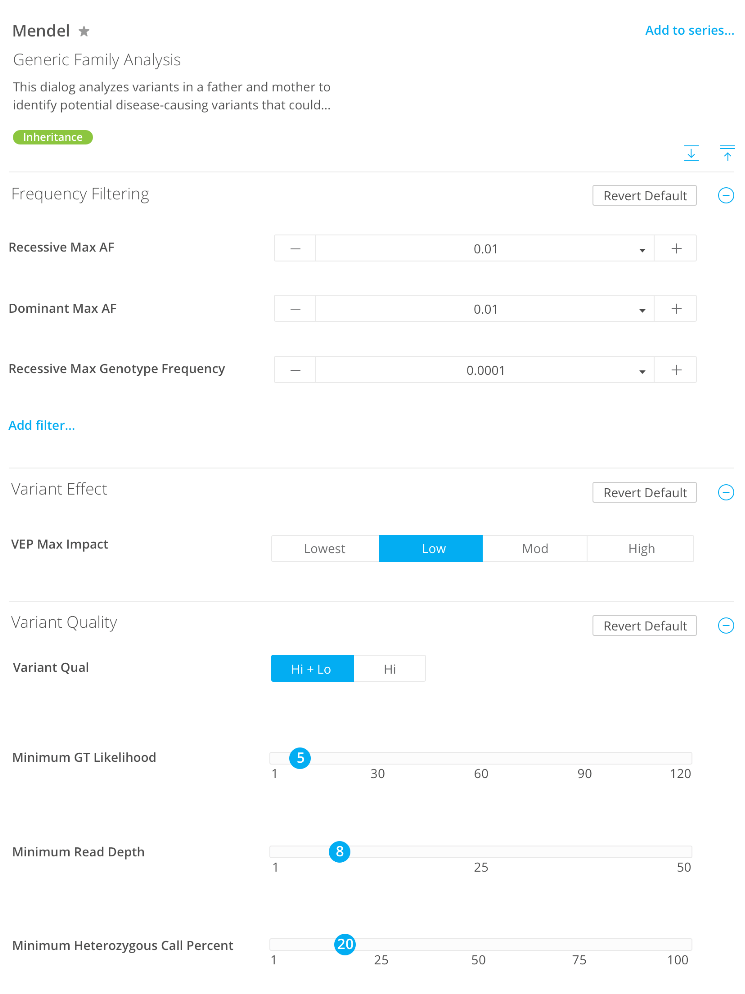
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Cases & Controls
Inheritance
Variants
Genes
Quality Control
Analysis Name
Description 1
Recommended for Selected Study



High
Variant Effect Predictor (VEP) Consequence


Transcript ablation


Splice acceptor variant


Splice donor variant


Stop gained


Frameshift variant


Stop lost


Start lost


Transcript amplification


Inframe insertion


Medium


Transcript ablation


Splice acceptor variant


Splice donor variant


Stop gained


Frameshift variant


Stop lost


Start lost


Transcript amplification


Inframe insertion
Genome range
Full Genome

High quality variants


Low quality variants


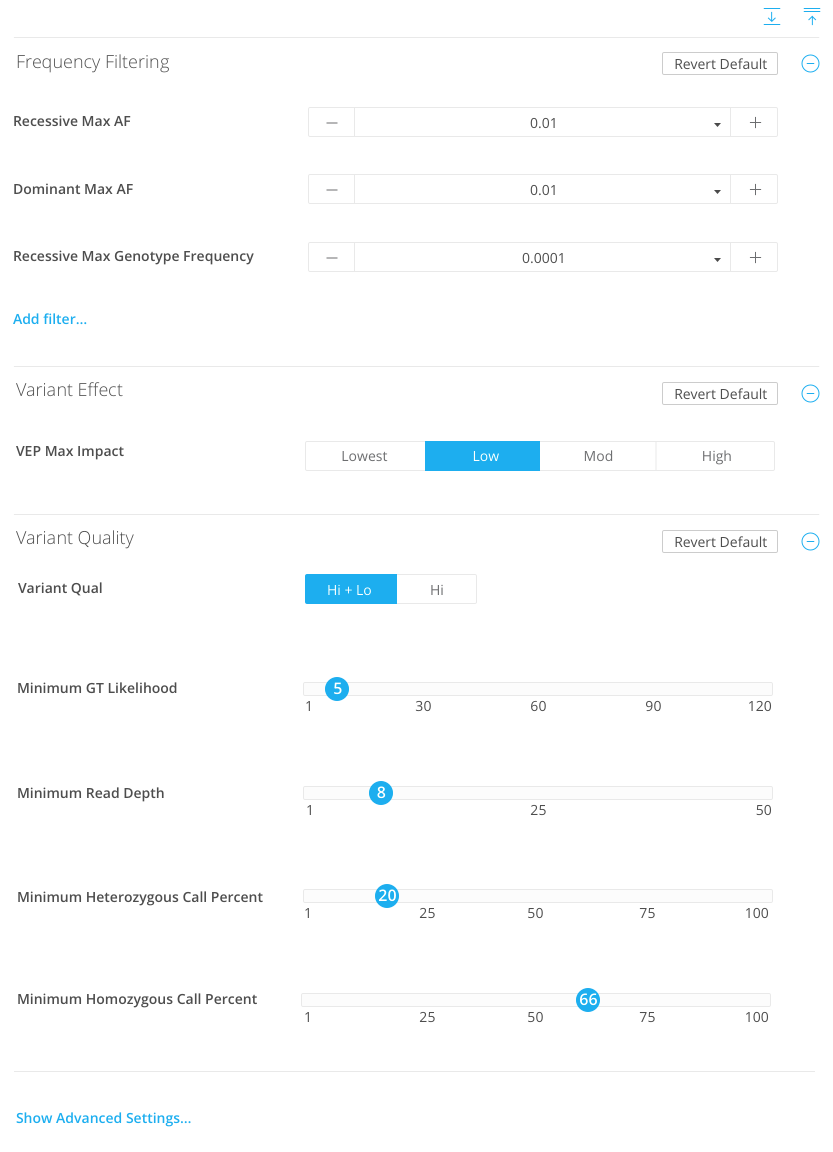
Quality
Minimum genotype likelihood
Minimum read depth
Minimum heterozygous call %
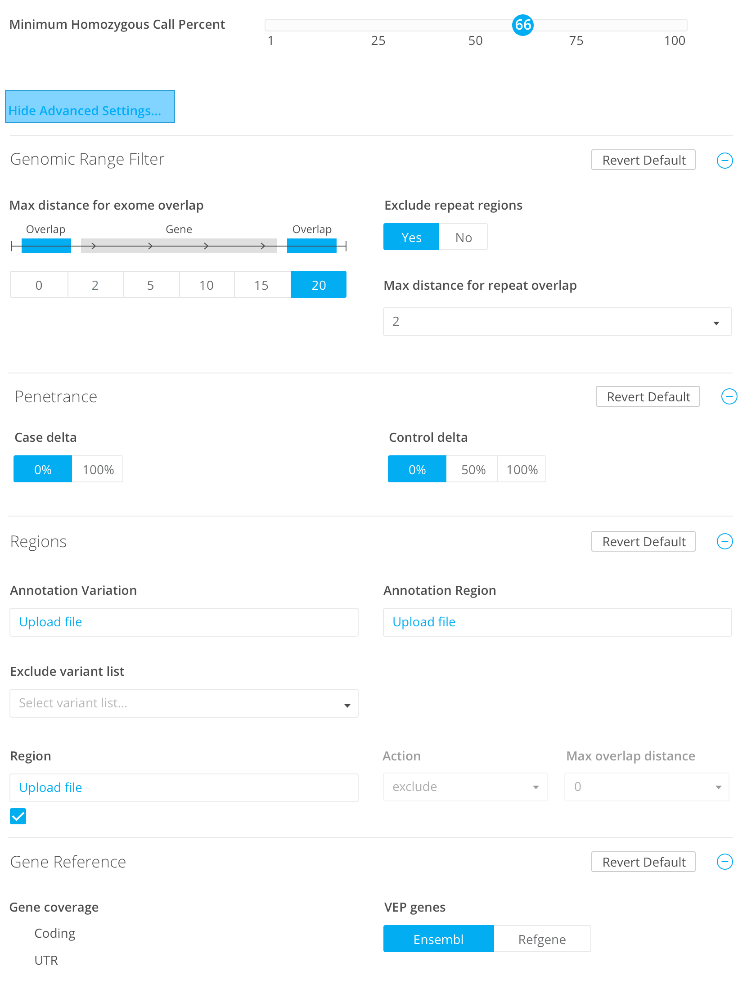
Minimum homozygous call %

Lowest

High

Low

Moderate
VEP Max Impact
Settings

Default

Frequency filtering








Variant quality

Varient effect

Genomic range filter

Penetrance
Case delta

0%

100%
Control delta

0%

50%

100%

Regions
Custom annotation variation

Upload file
Custom annotation region

Upload file
Custom region

Upload file
Exclude variants list
Select...

Default






Default






Default






Default






Default







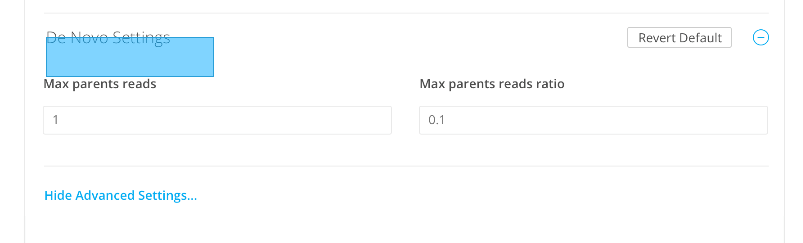
De novo settings
Default






Maximum parents reads
Maximum parents reads ratio
Enter your email...
Enter your email...


0

20

2

5

10

15
Maximum distance for exome overlap

Yes

No
Exclude repeat regions?
Maximum distance for repeat overlap?
2


Recessive Max AF
Dominant Max AF
Recessive Max Genotype Freq






0.1

0.05

0.04

0.03

0.02

0.01

0.005

0.002

0.001

0.0005







0.1

0.05

0.04

0.03

0.02

0.01

0.005

0.002

0.001

0.0005







0.1

0.05

0.04

0.03

0.02

0.01

0.005

0.002

0.001

0.0005


Gene reference
Default






Coding


UTR


Gene coverage
VEP genes

Ensembl

RefGene
Run analysis
Save as template
Analysis Name
Description








Apply template...

Selected item
Name
PN.UKBIOASC.PHYSMEA.ANTH.BODYSIZME.
Grand Total Count
499588
Statistics
Average
27.434210996664103
Maximum Value
74.6837
Minimum Value
12.1212
Standard Deviation
4.811708548798581
Generic Info
Path
PN/UK Biobank Assessment Centre/Physical Measures/Antropomethry/Body size measures
Add to Cohort >>
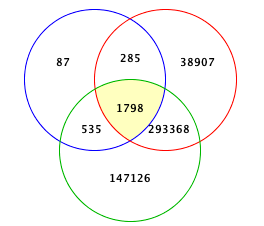
Venn
Query
Grid Builder

greater than

Focus on values
25
Add / drag elements to focus on
Cohort Info

Add or Drop Phenotypes here to see Venn diagram


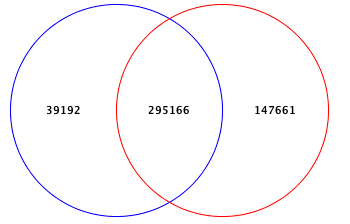

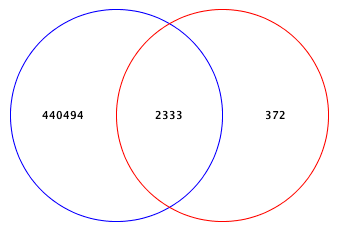
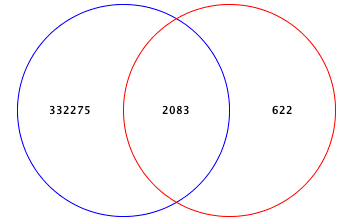
Name
Save Cohort...
Set to Grid
Untitled Cohort 1


Update automatically with new samples
Selected item
chr9
6216138
A
G
7.8E-6
40518
6821
286
0
304704
54147
2485
0
rs72614079
0.9437447786331177
0.94451504945755
0.8724383115768433
1.9999E-5
0.0270
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.947003
rs72614079
0.16780327260494232
chr9
6216138
A
G
7.8E-6
40518
6821
286
0
304704
54147
2485
0
rs72614079
0.9437447995205973
0.9445150354834543
0.8724382969482967
2.0E-5
0.027
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.947003
rs72614079
0.16780327815038296
chr9
6216240
C
G
0.01
28138
16925
2562
0
211300
130076
19960
0
rs1157505
0.9753394253022083
0.9792730137128878
0.9723698245302268
0.012
0.19
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.963959
rs1157505
0.45981843790328036
chr9
6216240
C
G
0.01
28138
16925
2562
0
211300
130076
19960
0
rs1157505
0.9753394253022083
0.9792730137128878
0.9723698245302268
0.012
0.19
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.963959
rs1157505
0.45981843790328036
chr9
6216516
C
G
1.7E-25
14203
23556
9866
0
100763
179750
80823
0
rs11791561
0.90996360787286
0.9303850061638009
0.9068566735954301
1.5E-18
2.1E-16
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.967862
rs11791561
0.933997933973316
chr9
6216516
C
G
1.7E-25
14203
23556
9866
0
100763
179750
80823
0
rs11791561
0.90996360787286
0.9303850061638009
0.9068566735954301
1.5E-18
2.1E-16
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.967862
rs11791561
0.933997933973316
chr9
6216524
C
G
0.0094
47284
339
2
0
358336
2990
9
0
rs185042275
0.8616961371016745
0.8646624794956047
1.686048058011185
0.0081
0.37
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.822078
rs185042275
0.008181601078355139
chr9
6216524
C
G
0.0094
47284
339
2
0
358336
2990
9
0
rs185042275
0.8616961371016745
0.8646624794956047
1.686048058011185
0.0081
0.37
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.822078
rs185042275
0.008181601078355139
chr9
6217849
G
T
6.6E-5
46110
1502
13
0
348580
12628
128
0
rs190777329
0.8978534703609695
0.8981724777130745
0.7705029719398471
7.7E-5
0.35
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.870772
rs190777329
0.03469416444915872
chr9
6217849
G
T
6.6E-5
46110
1502
13
0
348580
12628
128
0
rs190777329
0.8978534703609695
0.8981724777130745
0.7705029719398471
7.7E-5
0.35
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.870772
rs190777329
0.03469416444915872
chr9
6218346
G
A
1.3E-4
44264
3315
46
0
337550
23459
327
0
rs78121143
1.0775428587749776
1.0746769113562569
1.0673646077635695
1.1E-4
0.68
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.984564
rs78121143
0.06575723468380765
chr9
6218346
G
A
1.3E-4
44264
3315
46
0
337550
23459
327
0
rs78121143
1.0775428587749776
1.0746769113562569
1.0673646077635695
1.1E-4
0.68
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.984564
rs78121143
0.06575723468380765
chr9
6218595
C
T
4.1E-6
40853
6505
268
0
307159
51885
2292
0
rs72614080
0.939951432033209
0.9416902761549207
0.8864913476621246
7.7E-6
0.059
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.950057
rs72614080
0.1593190462219614
chr9
6218595
C
T
4.1E-6
40853
6505
268
0
307159
51885
2292
0
rs72614080
0.939951432033209
0.9416902761549207
0.8864913476621246
7.7E-6
0.059
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.950057
rs72614080
0.1593190462219614
chr9
6218711
CAT
C
1.1E-12
34900
11732
993
0
259374
93519
8442
0
9:6218711_CAT_C
0.9275230635546391
0.9330881637883944
0.8901493973043392
6.3E-12
5.0E-4
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.946491
9:6218711_CAT_C
0.29741286886372637
chr9
6218711
CAT
C
1.1E-12
34900
11732
993
0
259374
93519
8442
0
9:6218711_CAT_C
0.9275230635546391
0.9330881637883944
0.8901493973043392
6.3E-12
5.0E-4
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.946491
9:6218711_CAT_C
0.29741286886372637
chr9
6218960
T
A
1.3E-37
24954
19115
3556
0
199989
137766
23581
0
rs2066361
1.1260967892997091
1.1049119874642626
1.1557612384414893
5.8E-34
2.4E-14
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.977645
rs2066361
0.5185404003619137
chr9
6218960
T
A
1.3E-37
24954
19115
3556
0
199989
137766
23581
0
rs2066361
1.1260967892997091
1.1049119874642626
1.1557612384414893
5.8E-34
2.4E-14
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.977645
rs2066361
0.5185404003619137
chr9
6219010
C
T
2.6E-5
47567
58
0
0
361109
227
0
0
rs79203103
1.9397005420316478
1.939127987285053
1000.0
2.6E-5
1.0
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.959493
rs79203103
0.0018778705358333554
chr9
6219010
C
T
2.6E-5
47567
58
0
0
361109
227
0
0
rs79203103
1.9397005420316478
1.939127987285053
1000.0
2.6E-5
1.0
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.959493
rs79203103
0.0018778705358333554
chr9
6219176
G
T
5.2E-55
32415
13801
1409
0
258287
94341
8707
0
rs2066362
1.1761051965342093
1.1553801840601017
1.2347154658341368
4.4E-53
1.6E-12
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.971058
rs2066362
0.32481581382370867
chr9
6219176
G
T
5.2E-55
32415
13801
1409
0
258287
94341
8707
0
rs2066362
1.1761051965342093
1.1553801840601017
1.2347154658341368
4.4E-53
1.6E-12
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.971058
rs2066362
0.32481581382370867
chr9
6219845
A
C
5.3E-9
38994
8186
445
0
291954
65560
3822
0
rs1891385
0.931388656795901
0.9344179665631078
0.8822759579587534
1.5E-8
0.011
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.976937
rs1891385
0.20345876666208462
chr9
6219845
A
C
5.3E-9
38994
8186
445
0
291954
65560
3822
0
rs1891385
0.931388656795901
0.9344179665631078
0.8822759579587534
1.5E-8
0.011
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.976937
rs1891385
0.20345876666208462
chr9
6220750
ATTG
A
0.0073
47089
535
1
0
357752
3575
9
0
9:6220750_ATTG_A
1.1362120513435046
1.134707907427507
0.8430086604326484
0.0069
1.0
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.839798
9:6220750_ATTG_A
0.009366753588875053
chr9
6220750
ATTG
A
0.0073
47089
535
1
0
357752
3575
9
0
9:6220750_ATTG_A
1.1362120513435046
1.134707907427507
0.8430086604326484
0.0069
1.0
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.839798
9:6220750_ATTG_A
0.009366753588875053
chr9
6221162
C
G
5.5E-4
44611
2958
56
0
339862
21147
328
0
rs73396601
1.0692289912470363
1.0705908010155079
1.2957111338651068
9.4E-4
0.082
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.985847
rs73396601
0.06660867156741053
chr9
6221162
C
G
5.5E-4
44611
2958
56
0
339862
21147
328
0
rs73396601
1.0692289912470363
1.0705908010155079
1.2957111338651068
9.4E-4
0.082
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.985847
rs73396601
0.06660867156741053
chr9
6221246
T
C
5.3E-12
21854
20795
4976
0
160298
160716
40322
0
rs16924144
0.9402650227018489
0.9505702681625973
0.9288667712406045
3.2E-10
2.9E-6
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
1.0
rs16924144
0.6595569634536909
chr9
6221246
T
C
5.3E-12
21854
20795
4976
0
160298
160716
40322
0
rs16924144
0.9402650227018489
0.9505702681625973
0.9288667712406045
3.2E-10
2.9E-6
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
1.0
rs16924144
0.6595569634536909
chr9
6222110
T
C
8.1E-4
44593
2976
56
0
339690
21313
333
0
rs11794419
1.0670078494637587
1.0681819886115174
1.276234800252543
0.0013
0.1
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.985911
rs11794419
0.07013544066687319
chr9
6222110
T
C
8.1E-4
44593
2976
56
0
339690
21313
333
0
rs11794419
1.0670078494637587
1.0681819886115174
1.276234800252543
0.0013
0.1
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.985911
rs11794419
0.07013544066687319
chr9
6222149
T
C
1.1E-35
5501
21681
20443
0
37699
157963
165674
0
rs10815376
0.8919892546692109
0.9129914612446148
0.8882093713959919
1.7E-13
1.5E-33
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.990822
rs10815376
1.3486750880677212
chr9
6222149
T
C
1.1E-35
5501
21681
20443
0
37699
157963
165674
0
rs10815376
0.8919892546692109
0.9129914612446148
0.8882093713959919
1.7E-13
1.5E-33
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.990822
rs10815376
1.3486750880677212
chr9
6222553
G
A
0.001
44586
2983
56
0
339616
21381
339
0
rs11787939
1.0657626839730927
1.0666799224591972
1.2536257376414117
0.0016
0.13
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.984392
rs11787939
0.07381125092068469
chr9
6222553
G
A
0.001
44586
2983
56
0
339616
21381
339
0
rs11787939
1.0657626839730927
1.0666799224591972
1.2536257376414117
0.0016
0.13
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.984392
rs11787939
0.07381125092068469
chr9
6223806
C
G
4.4E-5
46865
755
4
0
354634
6665
38
0
rs191607042
0.8568490227141838
0.8576492744099826
0.7986449744689317
4.5E-5
0.81
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.924469
rs191607042
0.018029833261183115
chr9
6223806
C
G
4.4E-5
46865
755
4
0
354634
6665
38
0
rs191607042
0.8568490227141838
0.8576492744099826
0.7986449744689317
4.5E-5
0.81
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.924469
rs191607042
0.018029833261183115
chr9
6224308
T
C
6.1E-4
44608
2960
56
0
339834
21174
328
0
rs1929994
1.068578802919404
1.0699654102335487
1.295734783785779
0.001
0.082
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.987838
rs1929994
0.0667755622075095
chr9
6224308
T
C
6.1E-4
44608
2960
56
0
339834
21174
328
0
rs1929994
1.068578802919404
1.0699654102335487
1.295734783785779
0.001
0.082
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.987838
rs1929994
0.0667755622075095
chr9
6225659
G
A
3.0E-6
43852
3686
87
0
334799
26029
508
0
rs7048482
1.0854997492464824
1.086370769915739
1.2999147304617973
7.2E-6
0.029
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
1.0
rs7048482
0.07202164916938342
chr9
6225659
G
A
3.0E-6
43852
3686
87
0
334799
26029
508
0
rs7048482
1.0854997492464824
1.086370769915739
1.2999147304617973
7.2E-6
0.029
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
1.0
rs7048482
0.07202164916938342
chr9
6227045
G
C
3.9E-26
4758
20866
22001
0
32989
152369
175978
0
rs2006682
0.9051795871183567
0.9243807242064848
0.9043748003020572
1.6E-9
7.7E-25
IL33
ENST00000417746
+
.
.
5_prime_UTR_variant
LOWEST
435
4
0.99102
rs2006682
1.4019465028343752
chr9
6227045
G
C
3.9E-26
4758
20866
22001
0
32989
152369
175978
0
rs2006682
0.9051795871183567
0.9243807242064848
0.9043748003020572
1.6E-9
7.7E-25
IL33
ENST00000456383
+
.
.
5_prime_UTR_variant
LOWEST
687
6
0.99102
rs2006682
1.4019465028343752
Chrom
Position ▾
Reference
Call
pValue_mm
CASE_0_GTcount
ID
OR_dom
OR_mm
OR_rec
pval_dom
pval_rec
gene_symb
Transcript_stableid
strand
exonNum
exon
cdspos
Codons
abefore
aafter
Consequence
Impact
transcrsize
numExons
CASE_1_GTcount
CASE_2_GTcount
CASE_3_GTcount
CTRL_0_GTcount
CTRL_1_GTcount
CTRL_2_GTcount
CTRL_3_GTcount
information
rsID
AF
Drill in Reports ►
Web Links ►
Show transponded...
Open in new Grid
Open in new Grid as Cases
Open in new Grid as Controls
Copy
Copy with Header
Copy as Filter... ►
Copy as List... ►
Scroll to selected row
Invert selection
Duplicate last row





























































































































Run query


Column
Value
Name
Type
Group
The actual called sequence (variant), found by replacing a part of the reference sequence, denoted by Pos and Reference, with the sequence in the Call column.
Value
Row
Chrom
Pos
Ref
Call
->
Actions





Drill Ins ▼











